
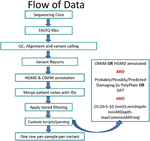
For more details on each argument, see the list further down below the table or click on an argument name to jump directly to that entry in the list. This table summarizes the command-line arguments that are specific to this tool. Similar to example 1 but we use an input list file to specify the input files: We combine several variant files in different formats, where at least one of them contains the contig list in its header. In a file with name ending in ".list" to INPUT.Ī VCF sorted (i) according to the dictionary and (ii) by coordiante. You can either directly specify the list of files by specifying INPUT multiple times, or provide a list Input files variant records must be sorted by their contig and position following the sequence dictionary provided.dict) if the input VCF does not contain aĬomplete contig list and if the output index is to be created (true by default). Optionally a sequence dictionary file (typically name ending in.Similar to example 1 but we use an input list file to specify the input files: java -jar picard.jar MergeVcfs \Ĭombines multiple variant files into a single variant file.
OutputsĪ VCF sorted (i) according to the dictionary and (ii) by coordinate. You can either directly specify the list of files by specifying INPUT multiple times, or provide a list in a file with name ending in ".list" to INPUT. Input files variant records must be sorted by their contig and position following the sequence dictionary provided or the header contig list.Input file headers must be contain compatible declarations for common annotations (INFO, FORMAT fields) and filters.If there are samples, those must be the same across all input files.The input variant data must adhere to the following rules: dict) if the input VCF does not contain a complete contig list and if the output index is to be created (true by default). ending in ".vcf.gz", or binary compressed, i.e. One or more input file in VCF format (can be gzipped, i.e.Combines multiple variant files into a single variant file.


 0 kommentar(er)
0 kommentar(er)
